Zooplankton and zoobenthos morphological parameters
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
-
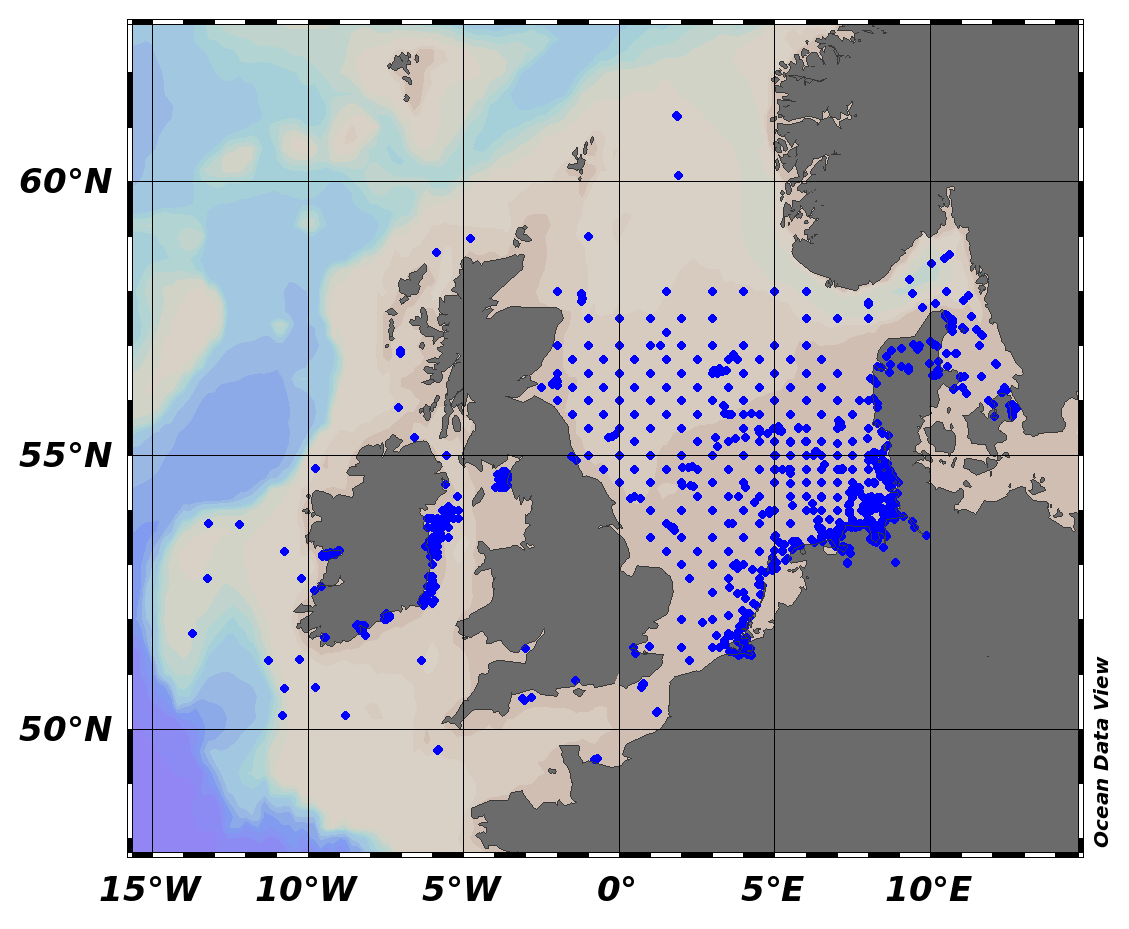
EMODnet Chemistry aims to provide access to marine chemistry data sets and derived data products concerning eutrophication, ocean acidification and contaminants. The chemicals chosen reflect importance to the Marine Strategy Framework Directive (MSFD). This regional aggregated dataset contains all unrestricted EMODnet Chemistry data on contaminants (49 parameters), and covers the Baltic Sea with 3818 CDI records divided per matrices: 1358 biota (396 Vertical profiles and 962 Time series),906 water profiles, 2510 sediment profiles. Vertical profiles temporal range is from 1985-04-16 to 2016-09-27. Time series temporal range is from 1972-05-02 to 2017-10-30. Data were aggregated and quality controlled by ‘Swedish Meteorological and Hydrological Institute (SMHI)’ from Sweden. Regional datasets concerning contaminants are automatically harvested. Parameter names in these datasets are based on P01, BODC Parameter Usage Vocabulary, which is available at: http://seadatanet.maris2.nl/bandit/browse_step.php . Each measurement value has a quality flag indicator. The resulting data collections for each Sea Basin are harmonised, and the collections are quality controlled by EMODnet Chemistry Regional Leaders using ODV Software and following a common methodology for all Sea Regions. Harmonisation means that: (1) unit conversion is carried out to express contaminant concentrations with a limited set of measurement units (according to EU directives 2013/39/UE; Comm. Dec. EU 2017/848) and (2) merging of variables described by different “local names” ,but corresponding exactly to the same concepts in BODC P01 vocabulary. The harmonised dataset can be downloaded as ODV spreadsheet (TXT file), which is composed of metadata header followed by tab separated values. This worksheet can be imported to ODV Software for visualisation (More information can be found at: https://www.seadatanet.org/Software/ODV ). The same dataset is offered also as XLSX file in a long/vertical format, in which each P01 measurement is a record line. Additionally, there are a series of columns that split P01 terms in subcomponents (measure, substance, CAS number, matrix...).This transposed format is more adapted to worksheet applications users (e.g. LibreOffice Calc). The 49 parameter names in this metadata record are based on P02, SeaDataNet Parameter Discovery Vocabulary, which is available at: http://seadatanet.maris2.nl/v_bodc_vocab_v2/vocab_relations.asp?lib=P02 . Detailed documentation will be published soon. The original datasets can be searched and downloaded from EMODnet Chemistry Download Service: https://emodnet-chemistry.maris.nl/search
-
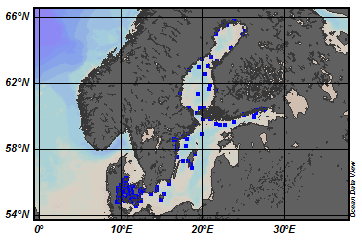
EMODnet Chemistry aims to provide access to marine chemistry data sets and derived data products concerning eutrophication, ocean acidification and contaminants. The chemicals chosen reflect importance to the Marine Strategy Framework Directive (MSFD). This regional aggregated dataset contains all unrestricted EMODnet Chemistry data on contaminants (20 parameters), and covers the North East Atlantic Ocean with 2400 CDI records divided per matrices: 122 in biota (as time series), 1689 in water (as vertical profiles), 589 in sediment (478 Vertical profiles and 111 Time series). Vertical profiles temporal range is from 1970-07-29 to 2017-02-28. Time series temporal range is from 1979-02-28 to 2014-10-21. Data were aggregated and quality controlled by ‘IFREMER / IDM / SISMER - Scientific Information Systems for the SEA’ from France. Regional datasets concerning contaminants are automatically harvested. Parameter names in these datasets are based on P01, BODC Parameter Usage Vocabulary, which is available at: http://seadatanet.maris2.nl/bandit/browse_step.php . Each measurement value has a quality flag indicator. The resulting data collections for each Sea Basin are harmonised, and the collections are quality controlled by EMODnet Chemistry Regional Leaders using ODV Software and following a common methodology for all Sea Regions. Harmonisation means that: (1) unit conversion is carried out to express contaminant concentrations with a limited set of measurement units (according to EU directives 2013/39/UE; Comm. Dec. EU 2017/848) and (2) merging of variables described by different “local names” ,but corresponding exactly to the same concepts in BODC P01 vocabulary. The harmonised dataset can be downloaded as ODV spreadsheet (TXT file), which is composed of metadata header followed by tab separated values. This worksheet can be imported to ODV Software for visualisation (More information can be found at: https://www.seadatanet.org/Software/ODV ). The same dataset is offered also as XLSX file in a long/vertical format, in which each P01 measurement is a record line. Additionally, there are a series of columns that split P01 terms in subcomponents (measure, substance, CAS number, matrix...).This transposed format is more adapted to worksheet applications users (e.g. LibreOffice Calc). The 20 parameter names in this metadata record are based on P02, SeaDataNet Parameter Discovery Vocabulary, which is available at: http://seadatanet.maris2.nl/v_bodc_vocab_v2/vocab_relations.asp?lib=P02 . Detailed documentation will be published soon. The original datasets can be searched and downloaded from EMODnet Chemistry Download Service: https://emodnet-chemistry.maris.nl/search
-
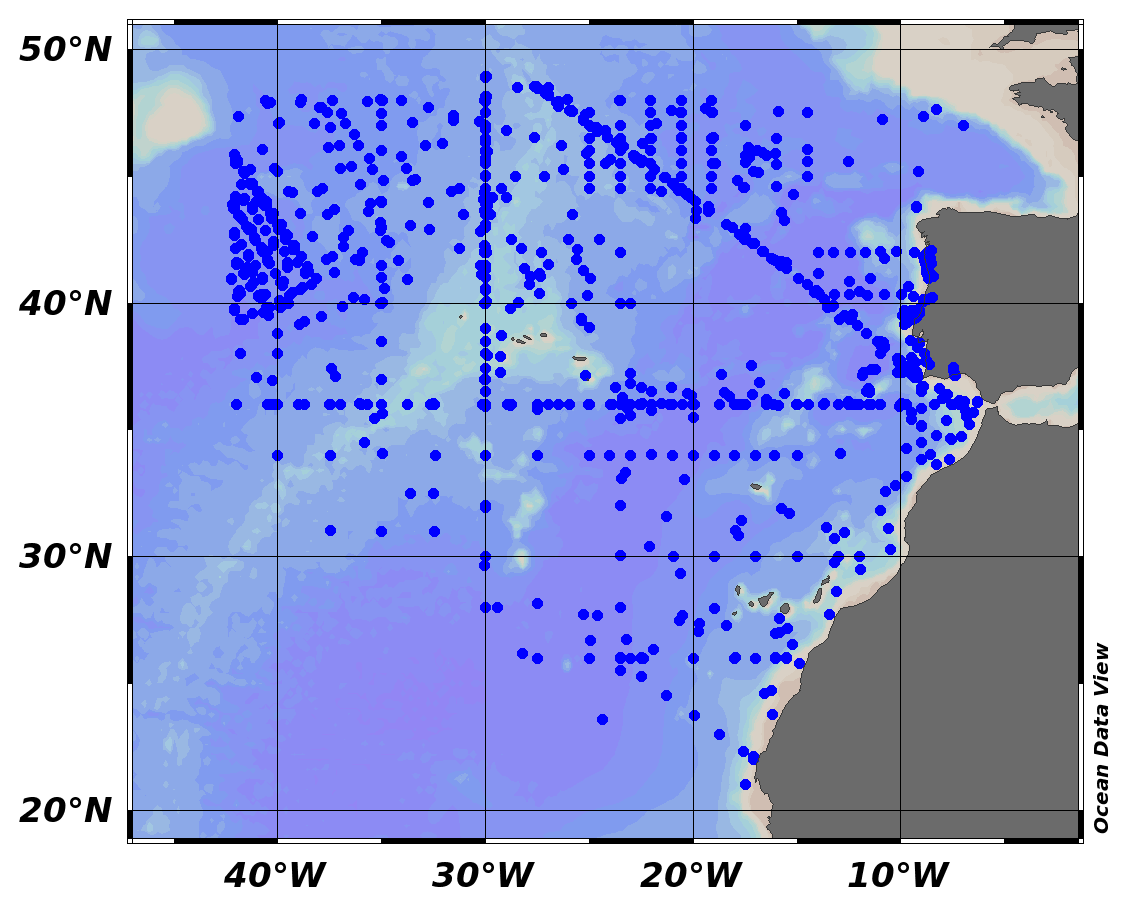
EMODnet Chemistry aims to provide access to marine chemistry data sets and derived data products concerning eutrophication, ocean acidification and contaminants. The chemicals chosen reflect importance to the Marine Strategy Framework Directive (MSFD). This regional aggregated dataset contains all unrestricted EMODnet Chemistry data on contaminants (59 parameters), and covers the North Sea with 34978 CDI records divided per matrices: 3909 biota time series, 28071 water profiles, 2998 sediment profiles. Vertical profiles temporal range is from 1970-02-17 to 2017-10-26. Time series temporal range is from 1979-02-26 to 2017-02-28. Data were aggregated and quality controlled by ‘Aarhus University, Department of Bioscience, Marine Ecology Roskilde from Denmark. Regional datasets concerning contaminants are automatically harvested. Parameter names in these datasets are based on P01, BODC Parameter Usage Vocabulary, which is available at: http://seadatanet.maris2.nl/bandit/browse_step.php . Each measurement value has a quality flag indicator. The resulting data collections for each Sea Basin are harmonised, and the collections are quality controlled by EMODnet Chemistry Regional Leaders using ODV Software and following a common methodology for all Sea Regions. Harmonisation means that: (1) unit conversion is carried out to express contaminant concentrations with a limited set of measurement units (according to EU directives 2013/39/UE; Comm. Dec. EU 2017/848) and (2) merging of variables described by different “local names” ,but corresponding exactly to the same concepts in BODC P01 vocabulary. The harmonised dataset can be downloaded as ODV spreadsheet (TXT file), which is composed of metadata header followed by tab separated values. This worksheet can be imported to ODV Software for visualisation (More information can be found at: https://www.seadatanet.org/Software/ODV ). The same dataset is offered also as XLSX file in a long/vertical format, in which each P01 measurement is a record line. Additionally, there are a series of columns that split P01 terms in subcomponents (measure, substance, CAS number, matrix...).This transposed format is more adapted to worksheet applications users (e.g. LibreOffice Calc). The 59 parameter names in this metadata record are based on P02, SeaDataNet Parameter Discovery Vocabulary, which is available at: http://seadatanet.maris2.nl/v_bodc_vocab_v2/vocab_relations.asp?lib=P02 . Detailed documentation will be published soon. The original datasets can be searched and downloaded from EMODnet Chemistry Download Service: https://emodnet-chemistry.maris.nl/search
-
Plankton was sampled with a Continuous Underway Fish Egg Sampler (CUFES, 315µm mesh size) at 4 m below the surface, and a WP2 net (200µm mesh size) from 100m to the surface, or 5 m above the sea floor to the surface when the depth was < 100 m, in the Bay of Biscay. The full images were processed with the ZooCAM software and the embedded Matrox Imaging Library (Colas et a., 2018) which generated regions of interest (ROIs) around each individual object and a set of features measured on the object. The same objects were re-processed to compute features with the scikit-image library http://scikit-image.org. The 1, 286, 590 resulting objects were sorted by a limited number of operators, following a common taxonomic guide, into 93 taxa, using the web application EcoTaxa http://ecotaxa.obs-vlfr.fr. For the purpose of training machine learning classifiers, the images in each class were split into training, validation, and test sets, with proportions 70%, 15% and 15%. The archive contains : taxa.csv.gz Table of the classification of each object in the dataset, with columns : objid : unique object identifier in EcoTaxa (integer number). taxon_level1 : taxonomic name corresponding to the level 1 classification lineage_level1 : taxonomic lineage corresponding to the level 1 classification taxon_level2 : name of the taxon corresponding to the level 2 classification plankton : if the object is a plankton or not (boolean) set : class of the image corresponding to the taxon (train : training, val : validation, or test) img_path : local path of the image corresponding to the taxon (of level 1), named according to the object id features_native.csv.gz Table of morphological features computed by ZooCAM. All features are computed on the object only, not the background. All area/length measures are in pixels. All grey levels are in encoded in 8 bits (0=black, 255=white). With columns : area : object's surface area_exc : object surface excluding white pixels area_based_diameter : object's Area Based Diameter: 2 * (object_area/pi)^(1/2) meangreyobjet : mean image grey level modegreyobjet : modal object grey level sigmagrey : object grey level standard deviation mingrey : minimum object grey level maxgrey : maximum object grey level sumgrey : object grey level integrated density: object_mean*object_area breadth : breadth of the object along the best fitting ellipsoid minor axis length : breadth of the object along the best fitting ellipsoid majorr axis elongation : elongation index: object_length/object_breadth perim : object's perimeter minferetdiam : minimum object's feret diameter maxferetdiam : maximum object's feret diameter meanferetdiam : average object's feret diameter feretelongation : elongation index: object_maxferetdiam/object_minferetdiam compactness : Isoperimetric quotient: the ration of the object's area to the area of a circle having the same perimeter intercept0, intercept45 , intercept90, intercept135 : the number of times that a transition from background to foreground occurs a the angle 0ø, 45ø, 90ø and 135ø for the entire object convexhullarea : area of the convex hull of the object convexhullfillratio : ratio object_area/convexhullarea convexperimeter : perimeter of the convex hull of the object n_number_of_runs : number of horizontal strings of consecutive foreground pixels in the object n_chained_pixels : number of chained pixels in the object n_convex_hull_points : number of summits of the object's convex hull polygon n_number_of_holes : number of holes (as closed white pixel area) in the object roughness : measure of small scale variations of amplitude in the object's grey levels rectangularity : ratio of the object's area over its best bounding rectangle's area skewness : skewness of the object's grey level distribution kurtosis : kurtosis of the object's grey level distribution fractal_box : fractal dimension of the object's perimeter hist25, hist50, hist75 : grey level value at quantile 0.25, 0.5 and 0.75 of the object's grey levels normalized cumulative histogram valhist25, valhist50, valhist75 : sum of grey levels at quantile 0.25, 0.5 and 0.75 of the object's grey levels normalized cumulative histogram nobj25, nobj50, nobj75 : number of objects after thresholding at the object_valhist25, object_valhist50 and object_valhist75 grey level symetrieh :index of horizontal symmetry symetriev : index of vertical symmetry skelarea : area of the object skeleton thick_r : maximum object's thickness/mean object's thickness cdist : distance between the mass and the grey level object's centroids features_skimage.csv.gz Table of morphological features recomputed with skimage.measure.regionprops on the ROIs produced by ZooCAM. See http://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops for documentation. inventory.tsv Tree view of the taxonomy and number of images in each taxon, displayed as text. With columns : lineage_level1 : taxonomic lineage corresponding to the level 1 classification taxon_level1 : name of the taxon corresponding to the level 1 classification n : number of objects in each taxon group map.png Map of the sampling locations, to give an idea of the diversity sampled in this dataset. imgs Directory containing images of each object, named according to the object id objid and sorted in subdirectories according to their taxon. Important Note: This submission has been initially submitted to SEA scieNtific Open data Edition (SEANOE) publication service and received the recorded DOI. The metadata elements have been further processed (refined) in EMODnet Ingestion Service in order to conform with the Data Submission Service specifications.
-
We report stable isotopes data, size measurements and environmental parameters for plankton collected between 2017 to 2019 in the Eastern English Channel and North Sea. Important Note: This submission has been initially submitted to SEA scieNtific Open data Edition (SEANOE) publication service and received the recorded DOI. The metadata elements have been further processed (refined) in EMODnet Ingestion Service in order to conform with the Data Submission Service specifications.
 EMODnet Product Catalogue
EMODnet Product Catalogue