features
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Scale 1:
-

-
-
-

-

-

Fluid emissions of non volcanic origin and mud-volcanoes on seafloors, mapped by various national and regional mapping projects and recovered in the literature. Polygons represent areas larger than 60 hectares. (for smaller areas, see fluid emissions points layer) Note: blank areas do not necessarily correspond to no occurrence.
-

Submarine landslides scarps, detected on the seabed, mapped by various national and regional mapping projects and recovered in the literature. Note: blank areas do not necessarily correspond to no occurrence.
-

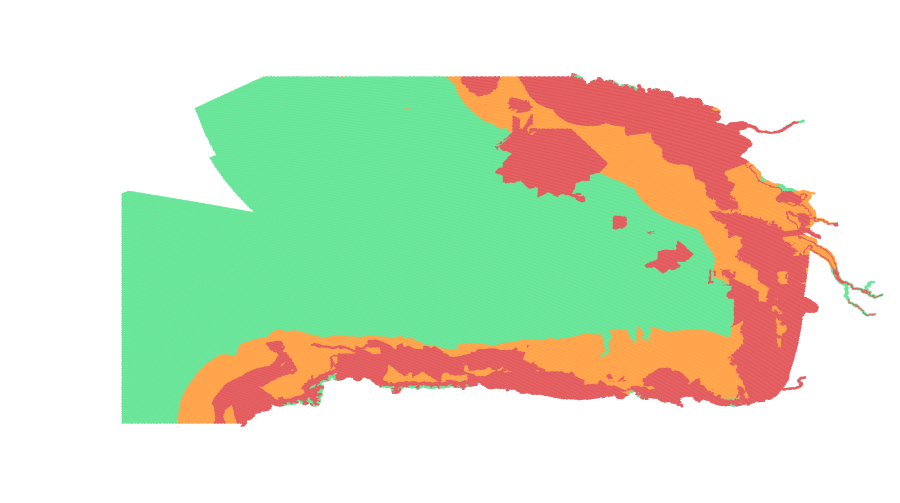
Submarine landslides, detectable on the seabed, outcropping or buried, mapped by various national and regional mapping projects and recovered in the literature. Locally landslides are extended on land to include their origin. Polygons represent areas larger than 60 hectares. Note: blank areas do not necessarily correspond to no occurrence.
-

Submarine landslides, detected on the seabed, outcropping or buried, mapped by various national and regional mapping projects and recovered in the literature. Locally polygons are extended on land to include the subaerial portions and/or origin of the landslides. Polygons represent areas larger than 10 hectares. (for smaller areas, see submarine landslides points layer) Note: blank areas do not necessarily correspond to no occurrence.
-

Submarine volcanoes, mapped by various national and regional mapping projects and recovered in the literature. Locally occurrences include their emerged portions. Polygons represent areas larger than 60 hectares. Note: blank areas do not necessarily correspond to no occurrence.
 EMODnet Product Catalogue
EMODnet Product Catalogue